Nonlinear Regression with ParmEst#
import sys
# If running on Google Colab, install Pyomo and Ipopt via IDAES
on_colab = "google.colab" in sys.modules
if on_colab:
!wget "https://raw.githubusercontent.com/dowlinglab/pyomo-doe/main/notebooks/tclab_pyomo.py"
# import TCLab model, simulation, and data analysis functions
from tclab_pyomo import (
TC_Lab_data,
TC_Lab_experiment,
extract_results,
extract_plot_results,
)
# set default number of states in the TCLab model
number_tclab_states = 2
Load and explore experimental data#
import pandas as pd
if on_colab:
file = "https://raw.githubusercontent.com/dowlinglab/pyomo-doe/main/data/tclab_sine_test_5min_period.csv"
else:
file = '../data/tclab_sine_test_5min_period.csv'
df = pd.read_csv(file)
df.head()
| Time | T1 | T2 | Q1 | Q2 | |
|---|---|---|---|---|---|
| 0 | 0.00 | 22.2 | 22.2 | 50 | 0 |
| 1 | 1.01 | 22.2 | 22.2 | 51 | 0 |
| 2 | 2.01 | 22.2 | 22.2 | 52 | 0 |
| 3 | 3.00 | 22.2 | 22.2 | 53 | 0 |
| 4 | 4.01 | 22.2 | 22.2 | 54 | 0 |
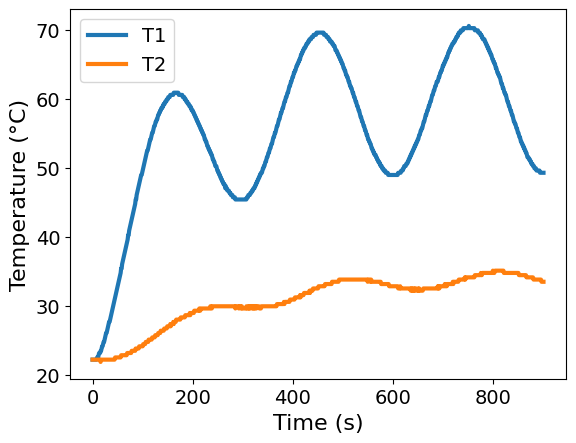
ax = df.plot(x='Time', y=['T1', 'T2'], xlabel='Time (s)', ylabel='Temperature (°C)')
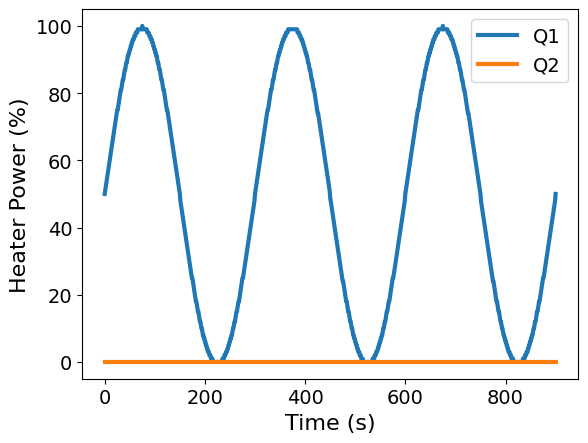
ax = df.plot(x='Time', y=['Q1', 'Q2'], xlabel='Time (s)', ylabel='Heater Power (%)')
Store in Custom Data Class#
In the file tclab_pyomo.py, we defined a dataclass for convenience. It is essentially a light weight container to store experimental data.
tc_data = TC_Lab_data(
name="Sine Wave Test for Heater 1",
time=df['Time'].values,
T1=df['T1'].values,
u1=df['Q1'].values,
P1=200,
TS1_data=None,
T2=df['T2'].values,
u2=df['Q2'].values,
P2=200,
TS2_data=None,
Tamb=df['T1'].values[0],
)
tc_data.to_data_frame().head()
| time | T1 | u1 | P1 | TS1_data | T2 | u2 | P2 | TS2_data | Tamb | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.00 | 22.2 | 50 | 200 | None | 22.2 | 0 | 200 | None | 22.2 |
| 1 | 1.01 | 22.2 | 51 | 200 | None | 22.2 | 0 | 200 | None | 22.2 |
| 2 | 2.01 | 22.2 | 52 | 200 | None | 22.2 | 0 | 200 | None | 22.2 |
| 3 | 3.00 | 22.2 | 53 | 200 | None | 22.2 | 0 | 200 | None | 22.2 |
| 4 | 4.01 | 22.2 | 54 | 200 | None | 22.2 | 0 | 200 | None | 22.2 |
Parameter Estimation with ParmEst#
We seek to solve minimize the sum of residuals squared:
subject to constraints
Here \(\bar{T}_{S,i}\) and \(\bar{u}_i\) are the measured sensor temperatures and control signals, respectives, at times \(i \in \mathcal{T}\). \(w\) is a small weight that helps regularize the solution by preventing the model predictions \(T_H\) from deviating too much from the measured temperatures.
In the tclab_pyomo.py model, we defined several helper functions:
extract_resultstakes a Pyomo model and returns the results stored in an instance of theTC_Lab_datadataclass.extract_plot_resultstakes experimental data (stored in aTC_Lab_datainstance) and a Pyomo model. The function then generates plots showing the data and model predictions.results_summarysummarizes the Pyomo.DoE results. We’ll use this later in the workshop.
parmest has been refactored to allow users to directly ask for the sum of squared error (SSE) objective. Custom objectives can also be provided, but are outside the scope of this workshop.
import pyomo.contrib.parmest.parmest as parmest
# First, we define an Experiment object within parmest
TC_Lab_sine_exp = TC_Lab_experiment(data=tc_data, number_of_states=number_tclab_states)
# Since everything has been labeled properly in the Experiment object, we simply invoke
# parmest's Estimator function to estimate the parameters.
pest = parmest.Estimator([TC_Lab_sine_exp, ], obj_function='SSE', tee=True)
obj, theta = pest.theta_est()
Ipopt 3.13.2:
******************************************************************************
This program contains Ipopt, a library for large-scale nonlinear optimization.
Ipopt is released as open source code under the Eclipse Public License (EPL).
For more information visit http://projects.coin-or.org/Ipopt
This version of Ipopt was compiled from source code available at
https://github.com/IDAES/Ipopt as part of the Institute for the Design of
Advanced Energy Systems Process Systems Engineering Framework (IDAES PSE
Framework) Copyright (c) 2018-2019. See https://github.com/IDAES/idaes-pse.
This version of Ipopt was compiled using HSL, a collection of Fortran codes
for large-scale scientific computation. All technical papers, sales and
publicity material resulting from use of the HSL codes within IPOPT must
contain the following acknowledgement:
HSL, a collection of Fortran codes for large-scale scientific
computation. See http://www.hsl.rl.ac.uk.
******************************************************************************
This is Ipopt version 3.13.2, running with linear solver ma27.
Number of nonzeros in equality constraint Jacobian...: 15301
Number of nonzeros in inequality constraint Jacobian.: 0
Number of nonzeros in Lagrangian Hessian.............: 7203
Total number of variables............................: 3606
variables with only lower bounds: 0
variables with lower and upper bounds: 1804
variables with only upper bounds: 0
Total number of equality constraints.................: 3602
Total number of inequality constraints...............: 0
inequality constraints with only lower bounds: 0
inequality constraints with lower and upper bounds: 0
inequality constraints with only upper bounds: 0
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
0 4.7112445e+04 3.12e-01 1.14e+01 -1.0 0.00e+00 - 0.00e+00 0.00e+00 0
1 1.6607309e+02 4.65e-02 1.98e+04 -1.0 9.60e+00 - 6.63e-01 1.00e+00f 1
2 1.7010095e+02 2.49e-03 1.41e+04 -1.0 9.30e-01 2.0 9.90e-01 1.00e+00h 1
3 5.4544219e+01 1.62e-04 3.53e+03 -1.0 5.34e-01 - 9.90e-01 1.00e+00f 1
4 5.4543738e+01 6.70e-07 4.99e-01 -1.0 8.96e-03 1.5 9.93e-01 1.00e+00h 1
5 5.3772867e+01 1.49e-04 1.35e+01 -1.7 9.12e-02 - 1.00e+00 1.00e+00f 1
6 5.3774283e+01 1.64e-08 1.21e-02 -1.7 1.08e-03 1.0 1.00e+00 1.00e+00h 1
7 5.3773994e+01 1.35e-07 7.20e-03 -3.8 3.27e-03 - 1.00e+00 1.00e+00h 1
8 5.3773993e+01 3.53e-14 1.03e-05 -3.8 2.79e-06 0.6 1.00e+00 1.00e+00h 1
9 5.3773993e+01 1.13e-05 4.97e-05 -5.7 3.41e-02 - 1.00e+00 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
10 5.3773955e+01 3.50e-01 1.10e+00 -5.7 5.90e+00 - 7.62e-05 7.38e-05H 1
11 5.3773961e+01 1.39e-02 5.68e+02 -5.7 2.93e+00 0.1 1.00e+00 1.00e+00h 1
12 5.3773979e+01 1.37e-02 5.59e+02 -5.7 5.63e-02 - 9.81e-01 1.56e-02h 7
13 5.3773987e+01 1.36e-02 5.55e+02 -5.7 5.61e-02 - 1.00e+00 7.81e-03h 8
14 5.3773991e+01 1.35e-02 5.53e+02 -5.7 5.57e-02 - 1.00e+00 3.91e-03h 9
15 5.3773992e+01 1.35e-02 5.52e+02 -5.7 5.54e-02 - 1.00e+00 9.77e-04h 11
16 5.3773992e+01 1.35e-02 5.52e+02 -5.7 5.54e-02 - 1.00e+00 2.44e-04h 13
17 5.3775014e+01 1.42e-04 1.13e+01 -5.7 5.54e-02 - 1.00e+00 1.00e+00h 1
18 5.3773992e+01 1.49e-08 2.46e-03 -5.7 5.74e-04 -0.4 1.00e+00 1.00e+00h 1
19 5.3773993e+01 8.04e-13 1.49e-06 -5.7 1.09e-05 -0.9 1.00e+00 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
20 5.3773993e+01 2.25e-07 7.08e-07 -8.6 2.47e-03 - 1.00e+00 1.00e+00h 1
21 5.3773993e+01 3.85e-02 1.17e-01 -8.6 1.03e+00 - 1.00e+00 1.00e+00h 1
22 5.3775641e+01 1.16e-03 3.45e-01 -8.6 3.08e-01 -1.3 1.00e+00 1.00e+00s 22
23 5.3774040e+01 9.24e-07 6.50e-03 -8.6 7.08e-03 - 1.00e+00 1.00e+00s 22
24 5.3773993e+01 3.04e-11 1.35e-03 -8.6 3.57e-05 - 1.00e+00 0.00e+00S 22
25 5.3773993e+01 8.45e-10 5.87e-08 -8.6 2.17e-04 - 1.00e+00 1.00e+00h 1
26 5.3773993e+01 1.27e-02 3.85e-02 -8.6 3.39e+00 - 1.00e+00 2.50e-01h 3
27 5.3774047e+01 1.17e-04 6.38e-03 -8.6 6.78e-02 -1.8 1.00e+00 1.00e+00s 22
28 5.3773993e+01 9.22e-09 6.67e-05 -8.6 8.53e-04 - 1.00e+00 1.00e+00s 22
29 5.3773993e+01 2.53e-07 1.32e-04 -8.6 4.53e-03 - 1.00e+00 0.00e+00S 22
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
30 5.3773993e+01 9.69e-04 2.92e-03 -8.6 2.81e-01 - 1.00e+00 1.00e+00h 1
31 5.3773993e+01 9.69e-04 2.92e-03 -8.6 4.50e-03 -2.3 1.00e+00 2.44e-04h 13
32 5.3773993e+01 9.60e-04 2.90e-03 -8.6 3.03e-01 - 1.00e+00 1.00e+00h 1
33 5.3773993e+01 8.40e-04 2.53e-03 -8.6 3.96e-03 -2.8 1.00e+00 1.25e-01h 4
34 5.3773993e+01 6.30e-04 1.90e-03 -8.6 6.84e-03 - 1.00e+00 2.50e-01h 3
35 5.3773993e+01 4.73e-04 1.42e-03 -8.6 5.50e-03 - 1.00e+00 2.50e-01h 3
36 5.3773993e+01 3.54e-04 1.07e-03 -8.6 4.60e-03 - 1.00e+00 2.50e-01h 3
37 5.3773993e+01 2.66e-04 8.01e-04 -8.6 4.08e-03 - 1.00e+00 2.50e-01h 3
38 5.3773993e+01 2.16e-08 1.12e-07 -8.6 3.90e-03 - 1.00e+00 1.00e+00h 1
39 5.3773993e+01 2.72e-14 9.17e-11 -8.6 1.62e-07 -3.2 1.00e+00 1.00e+00h 1
Number of Iterations....: 39
(scaled) (unscaled)
Objective...............: 5.3773992845814625e+01 5.3773992845814625e+01
Dual infeasibility......: 9.1707767478106112e-11 9.1707767478106112e-11
Constraint violation....: 2.7172708527700706e-14 2.7172708527700706e-14
Complementarity.........: 2.5059035596800667e-09 2.5059035596800667e-09
Overall NLP error.......: 2.5059035596800667e-09 2.5059035596800667e-09
Number of objective function evaluations = 154
Number of objective gradient evaluations = 40
Number of equality constraint evaluations = 154
Number of inequality constraint evaluations = 0
Number of equality constraint Jacobian evaluations = 40
Number of inequality constraint Jacobian evaluations = 0
Number of Lagrangian Hessian evaluations = 39
Total CPU secs in IPOPT (w/o function evaluations) = 0.285
Total CPU secs in NLP function evaluations = 0.071
EXIT: Optimal Solution Found.
parmest_regression_results = extract_plot_results(
tc_data, pest.ef_instance
)
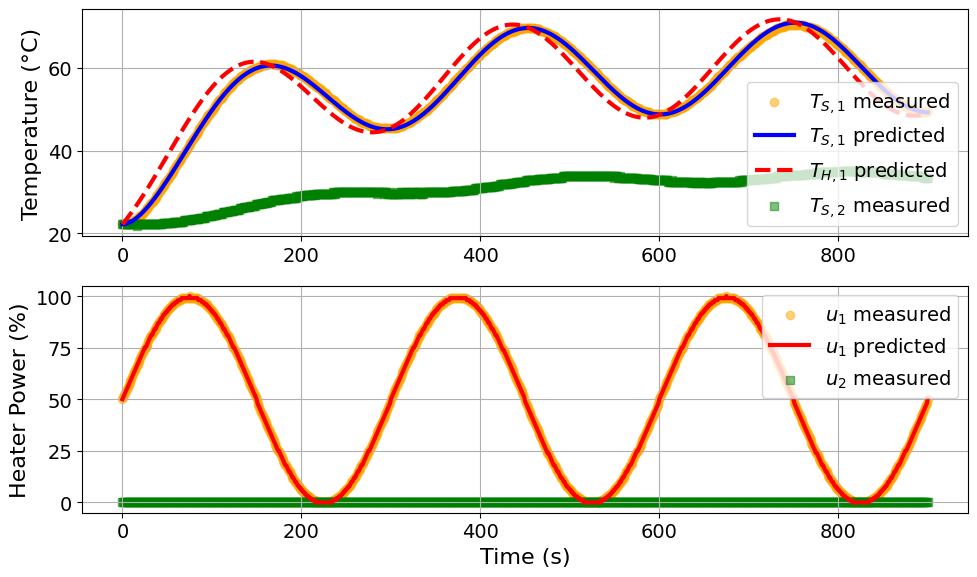
Model parameters:
Ua = 0.0417 Watts/degC
Ub = 0.0094 Watts/degC
CpH = 6.0274 Joules/degC
CpS = 0.1714 Joules/degC
Let’s see how to access the regression results:
theta_values = theta.to_dict()
print("Estimated parameters:\n", theta_values)
Estimated parameters:
{'Ub': 0.009440714239773074, 'inv_CpS': 5.8357556063605465, 'Ua': 0.0417051733576387, 'inv_CpH': 0.1659093525658045}

